Initially, ARB was developed for rRNA data, however, the package also supports handling and analysis of protein coding genes.
Highlighted features_
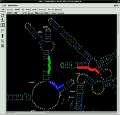
- Any database entry can be visualised in the main window along with a phylogenetic tree. Database access and navigation is possible via mouse click in the displayed tree or by using a search tool.
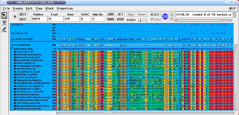
- Sequence and additional data can be im- and exported in a variety of commonly used flat file formates.
- Publication ready trees can be generated and exported in various formates.
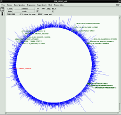
- Distance matrix, maximum parsimony and maximum likelihood based phylogenetic treeing can be performed applying the respective integrated tools. A special maximum parsimony approach allows reconstruction and optimisation of comprehensive trees representing the full sequence data set (currently more than 20.000 entries).
- A powerful editor for real or virtual primary structures includes versatile tools for string searching, automated alignment of primary structures, local alignment optimisation, automated secondary structure check as well as secondary structure visualisation.
- Conservation profiles and column filters can be established according to user defined criteria applying different procedures.
- The ARB PT server (positional tree) provides the basis for rapid searching closest relatives or specific sequence signatures. Such signatures can be evaluated as taxon specific probes against the background of the full database.
- The ARB project maintains processed databases for ribosomal RNAs and selected evolutionary conserved genes.
- ARB can as well handle and visualize annotated genome sequence data. To activate that feature, import annotated data from EMBL or Genbank.
- More about 'The ARB-Project' (PDF; 1.9 Mb)
- Last Update : 2003-Feb-20